Tag: protein design
-
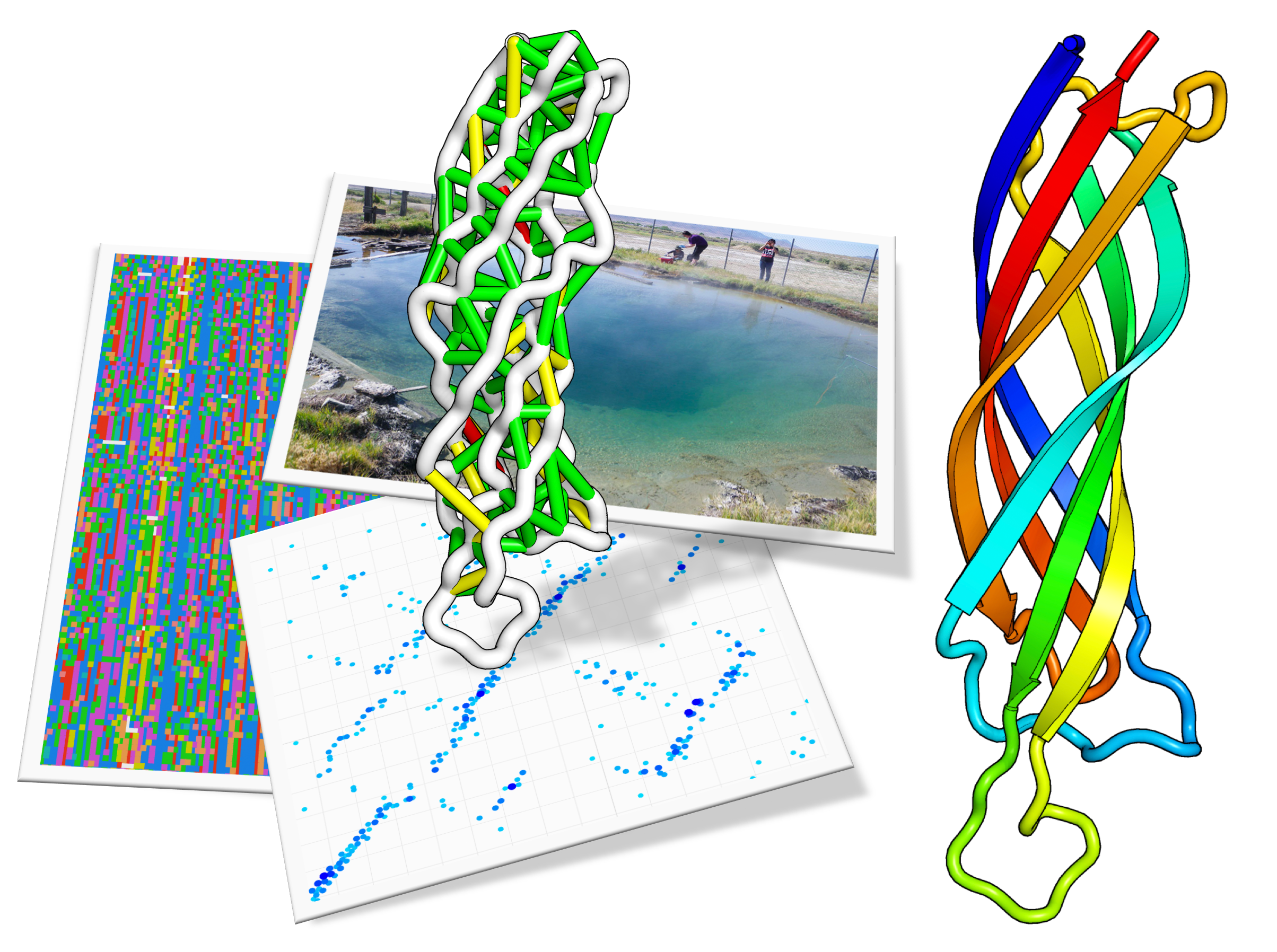
Protein structure determination using metagenome sequence data
Despite decades of work by structural biologists, there are still ~5200 protein families with unknown structure outside the range of comparative modeling. We show that Rosetta structure prediction guided by residue-residue contacts inferred from evolutionary information can accurately model proteins that belong to large families and that metagenome sequence data more than triple the number…
-
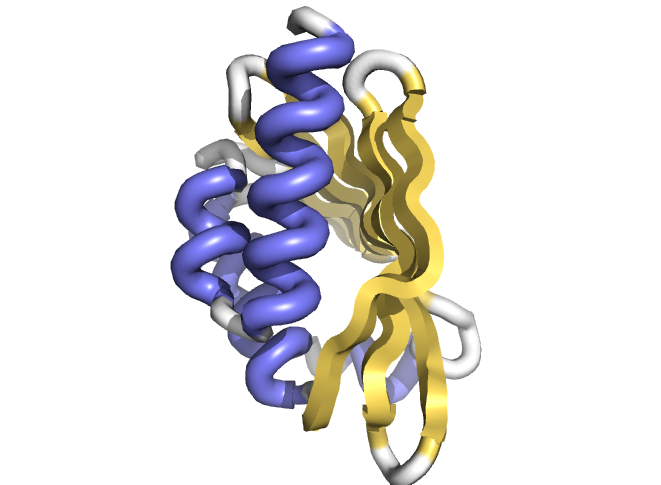
Principles for designing proteins with cavities formed by curved β sheets
Some of the key functions of the proteins in our bodies and in all living things are to catalyze chemical reactions—speed up the rates by many orders of magnitude-and to sense and respond to small molecules in the body and in the environment. New proteins that catalyze chemical reactions and/or sense and respond to compounds…
-
Baker Lab website soon to be searchable on Google!
For some reason, this website was not populating when any combination of “Baker” and “lab” was searched through Google. We hope to have remedied this, and I am using this post as a way to get Google to crawl faster (by adding new “content”). Thanks to everyone who has been notifying me of this issue.…
-

Halloween 2016 – Rise of the Davids
Happy Halloween! We celebrated early because a party on Monday isn’t as good as a Friday, of course. Take a look at the great costumes this year – it’ll have you seeing double, no, a score! There was also a menagerie of animals running amok, a beaten-up pinata, and an absolutely TERRIFYING scarecrow. [envira-gallery id=”1481″]
-
Accurate de novo design of hyperstable constrained peptides
Small constrained peptides combine the stability of small molecule drugs with the selectivity and potency of antibody-based therapeutics. However, peptide-based therapeutics have largely remained underexplored due to the limited diversity of naturally occurring peptide scaffolds, and a lack of methods to design them rationally. In an article published in Nature this week, Baker lab scientists…
-
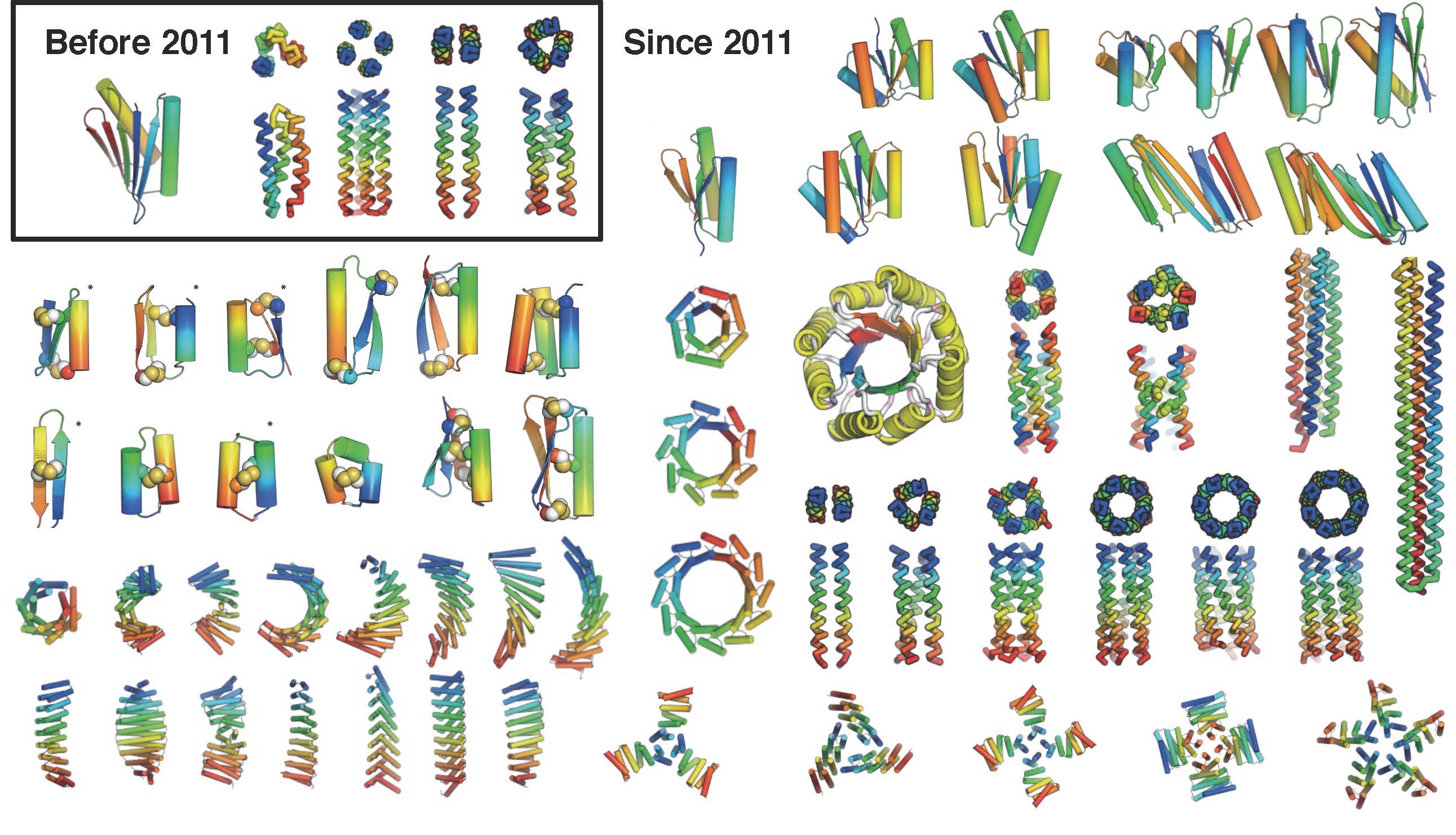
The coming of age of de novo protein design
Most protein design efforts to date have focused on reengineering existing proteins found in nature. By contrast, de novo protein design generates new structures from scratch, with sequences unrelated to naturally occurring proteins. Before 2011, the only successful de novo designed proteins were Top7 (2003), and an array of coiled coil peptides (helical bundles). In…
-
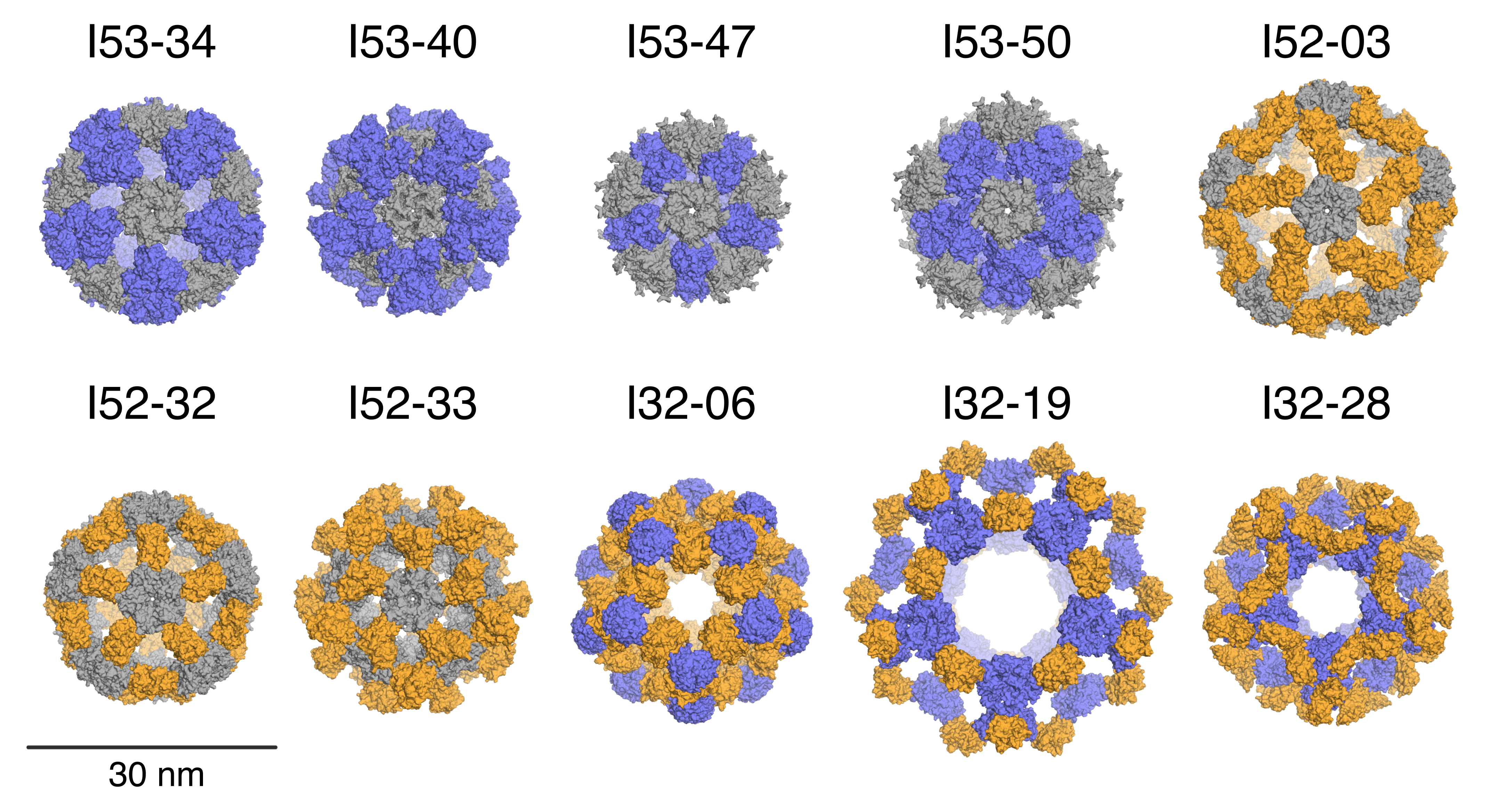
Accurate design of megadalton-scale two-component icosahedral protein complexes
Nature provides many examples of self- and co-assembling protein-based molecular machines, including icosahedral protein cages that serve as scaffolds, enzymes, and compartments for essential biochemical reactions and icosahedral virus capsids, which encapsidate and protect viral genomes and mediate entry into host cells. Inspired by these natural materials, we report the computational design and experimental characterization…
-
Arrivederci, Fabio!
This past Friday we bid a fond farewell to Fabio Parmegianni as he embarked on his next stage of life, or as some call it, the P.B. (Post-Bakerite) era. Though we will miss him, we wish him the best of luck at the University of Bristol in the UK. The lab will undoubtedly be a…