Tag: binder
-
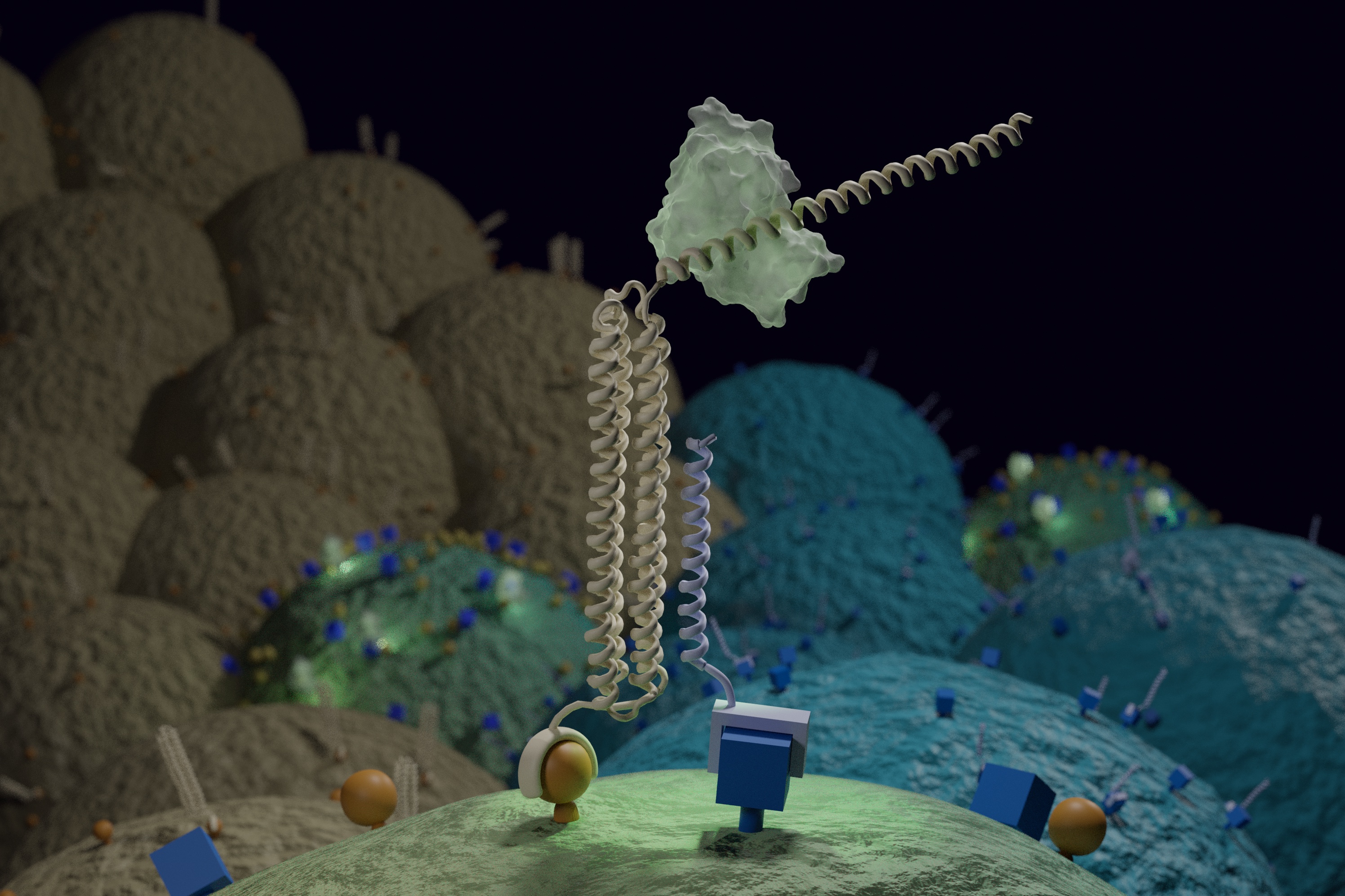
Introducing Co-LOCKR: designed protein logic for cell targeting
In a new paper [PDF] appearing in Science, a team of IPD researchers together with colleagues at UW Medicine and Fred Hutchinson Cancer Research Center demonstrate a new way to precisely target cells — including those that look almost exactly like their neighbors. They designed nanoscale devices made of synthetic proteins that target a therapeutic agent only to…
-
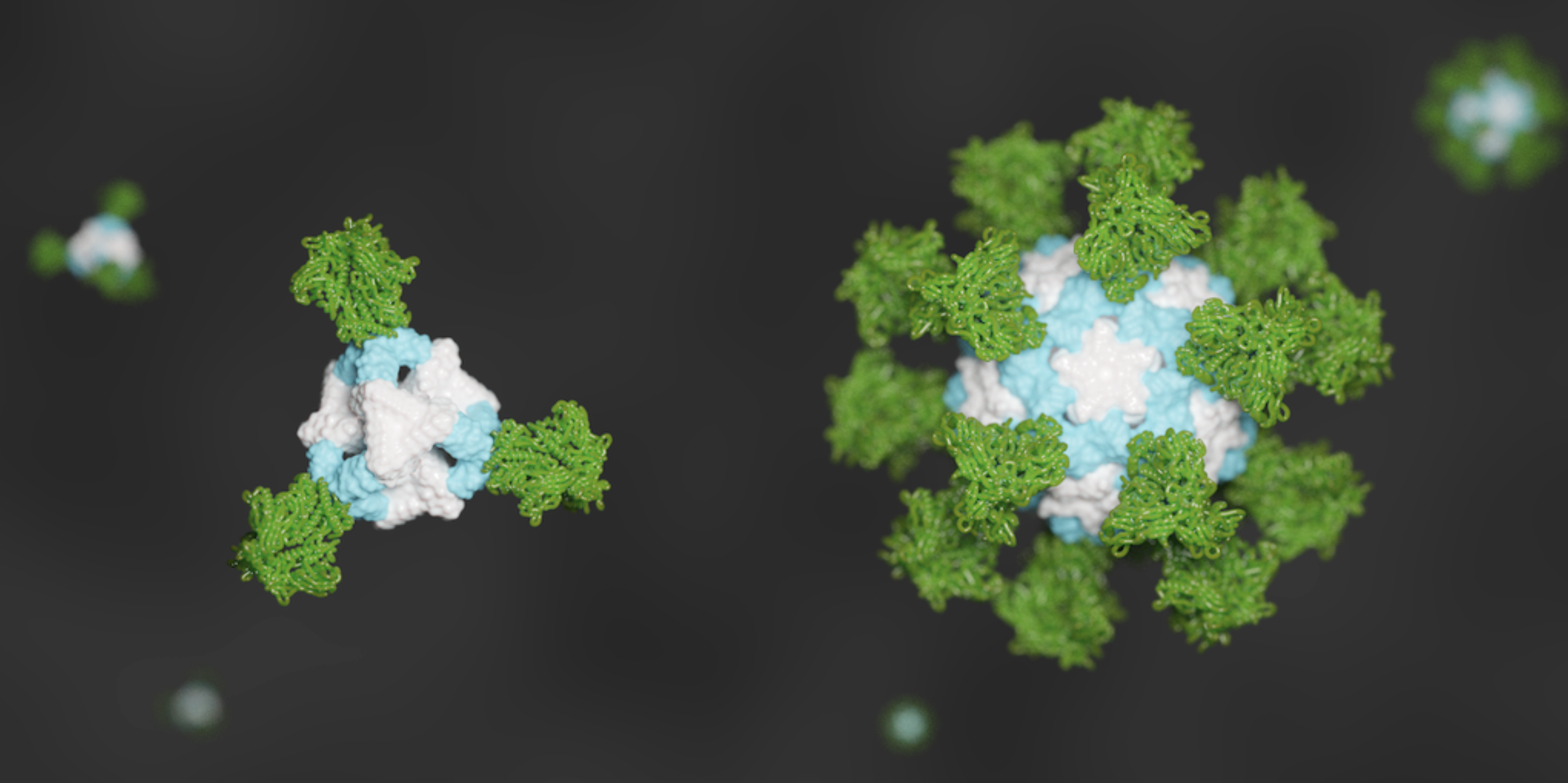
De novo nanoparticles as vaccine scaffolds
IPD researchers have developed a new vaccine design strategy that could confer improved immunity against certain viruses, including those that cause AIDS, the flu, and COVID-19. Using this technique, viral antigens are attached to the surface of self-assembling, de novo designed protein nanoparticles. This enables an unprecedented level of control over the molecular configuration of the resulting…
-
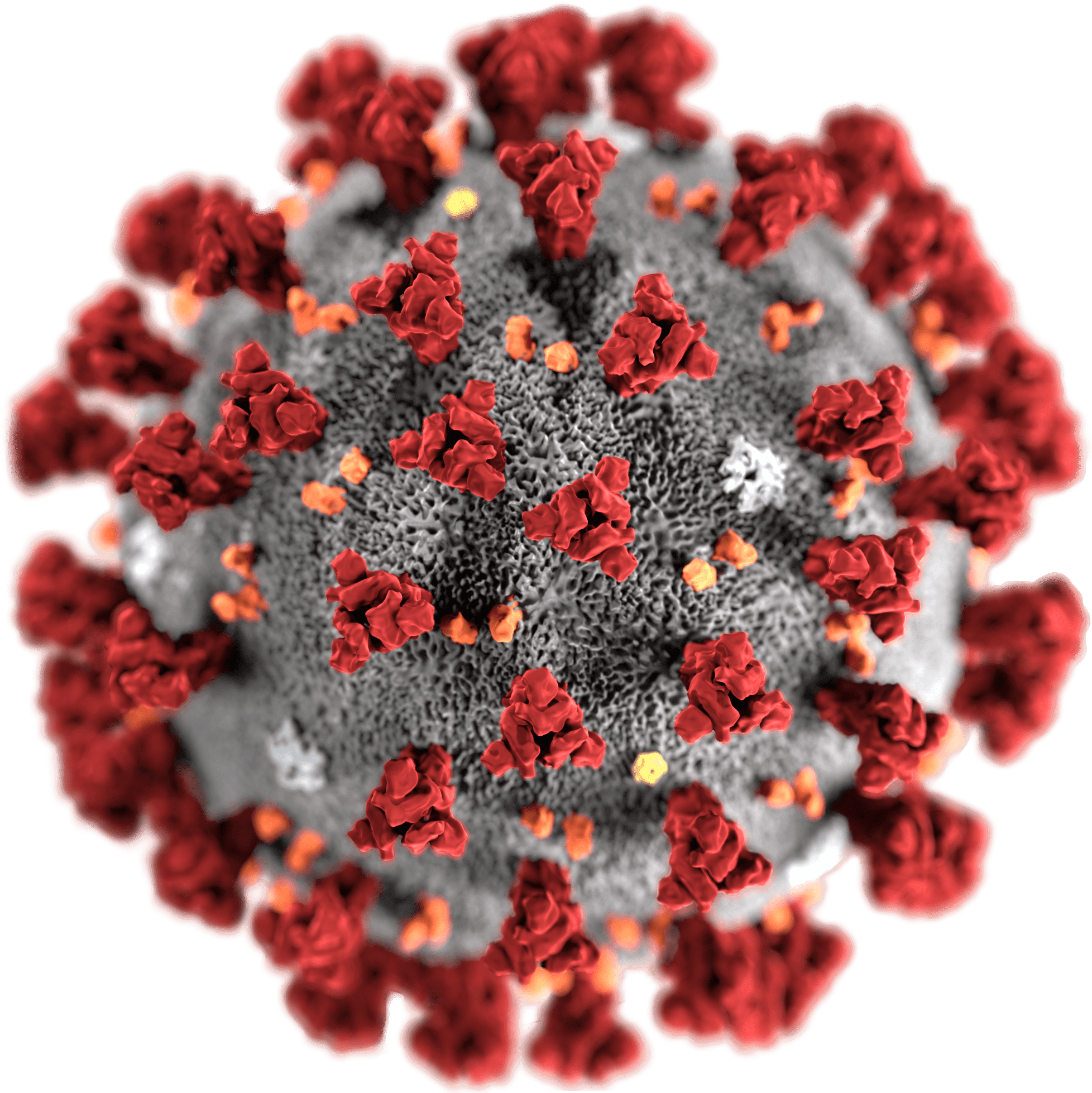
Rosetta’s role in fighting coronavirus
To follow updates on our COVID-19 research, visit ipd.uw.edu/coronavirus
-
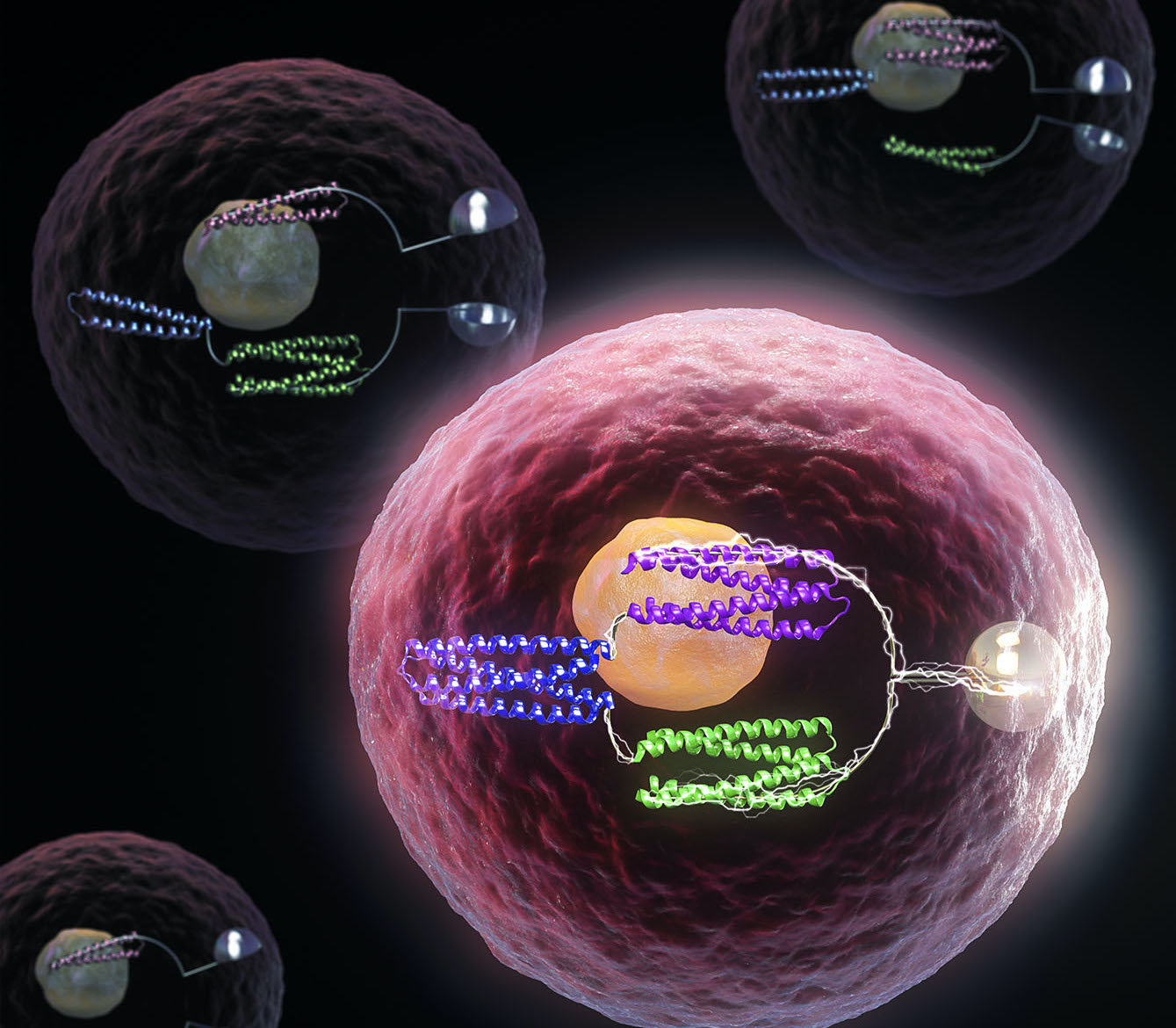
De novo design of protein logic gates
The same basic tools that allow computers to function are now being used to control life at the molecular level, with implications for future medicines and synthetic biology. Together with collaborators, we have created artificial proteins that function as molecular logic gates. These tools, like their electronic counterparts, can be used to program the behavior of…
-
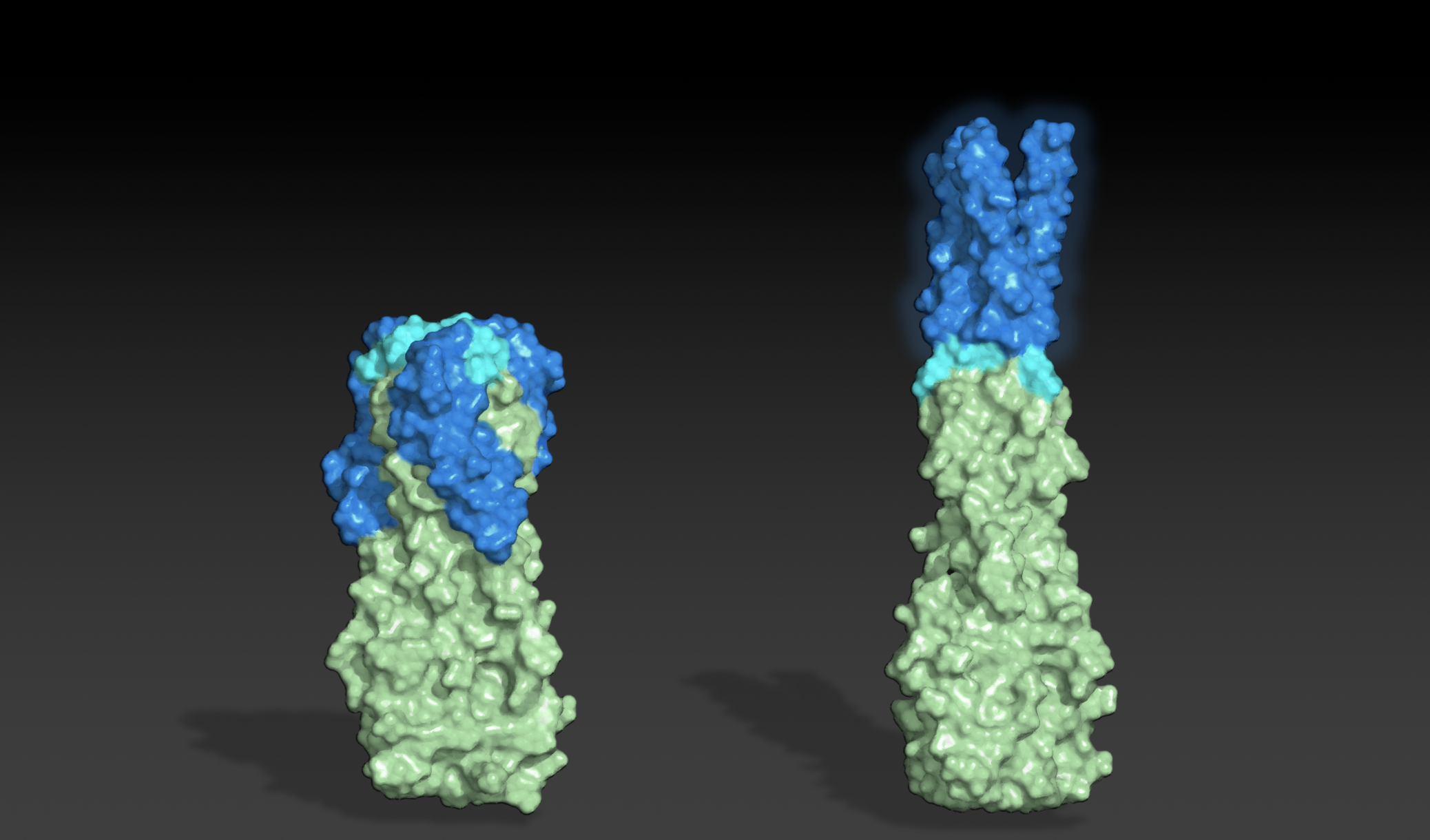
Designing shape-shifting proteins
Today we report the design of protein sequences that adopt more than one well-folded structure, reminiscent of viral fusion proteins. This research moves us closer to creating artificial protein systems with reliable moving parts. In nature, many proteins change shape in response to their environment. This plasticity is often linked to biological function. While computational…
-

SCI-STEM Symposium 2020
The Institute for Protein Design at the University of Washington held the second symposium aimed at providing strategies to address diversity challenges in science, technology, engineering, and math (STEM). The Strategies for Cultivating Inclusion in STEM (SCI-STEM) symposium featured leading keynote speakers, panel discussions, and interactive breakout sessions. Members of the STEM community at all…
-

Congrats, Dr Nattermann!
Baker lab graduate student Una Nattermann defended today! Her talk titled “A Hierarchical Approach To The Design Of Protein Crystals” was attended by friends and family as well as current and former members of the lab.
-

Neville wins HHMI Hanna Gray Fellowship!
Congratulations to postdoc Neville Bethel for his selection as a Hanna H. Gray Fellow! Neville is one of 15 exceptional early career scientists selected by HHMI for support this year. His research is focused on understand through design the mechanical properties of supramolecular assemblies. “HHMI is committed to supporting people who will solve some of the greatest problems in science,”…