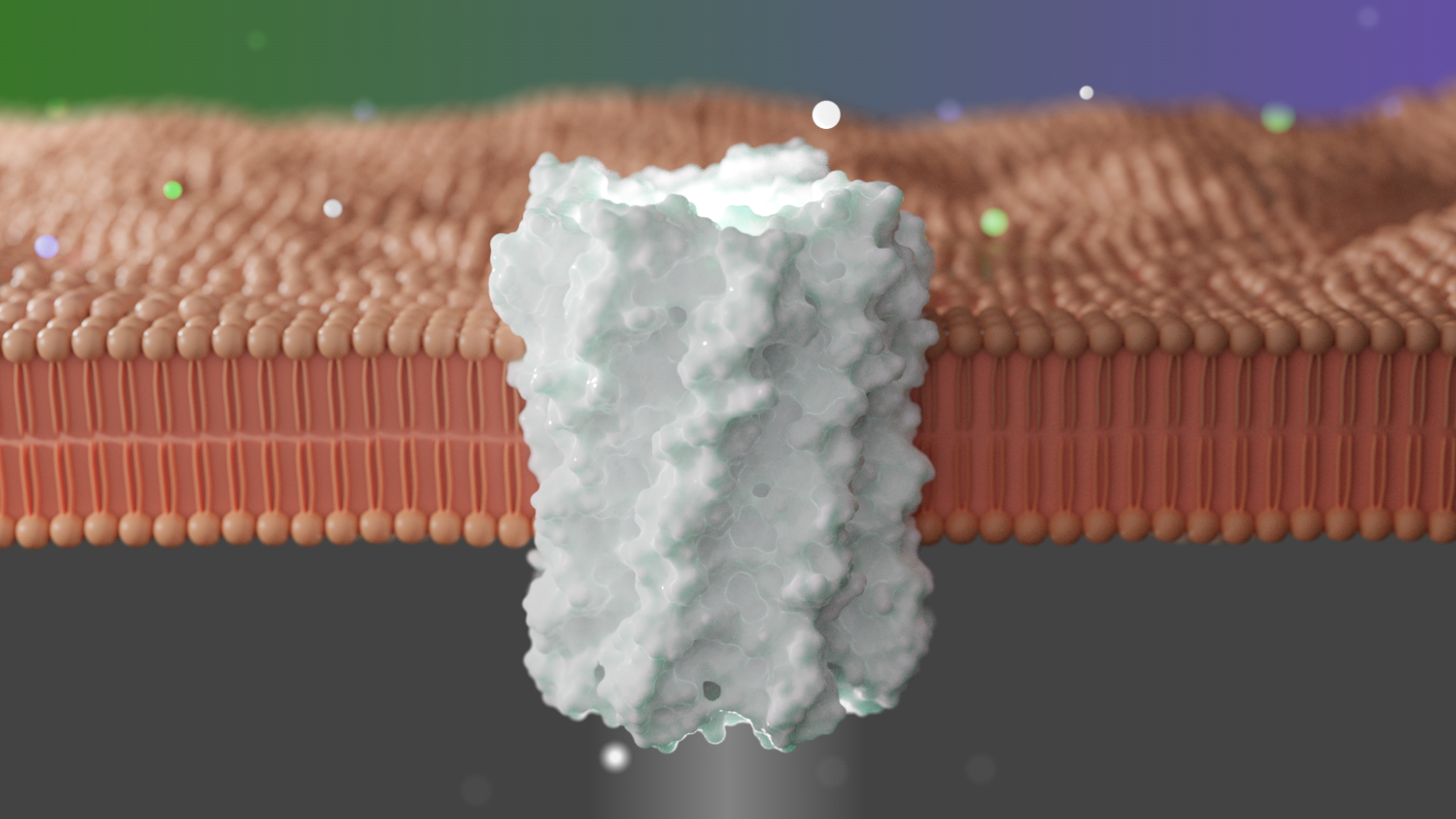
This week we report in Nature [PDF] the design of new transmembrane proteins that allow cells to take in certain chemicals, including charged ions and larger fluorescent molecules. This research could enable new forms of drug delivery and allow for better control over the electrical activity of living cells.
All cells are studded with tiny protein pores that take in and expel ions such as sodium and potassium. This is essential for life and enables the electrical activity of the brain. Biochemists have for decades studied how ion channel proteins work, but creating new ones has proven extremely challenging.
A team of researchers including biochemists from the University of Washington, Westlake University, the University of Cambridge, and Osaka University designed and characterized new pores formed by two concentric rings of alpha-helices that are stable and monodisperse. The lead authors of this work are Chunfu Xu and Peilong Lu, current and former Baker lab postdoctoral scholars, respectively.
“We decided to use a two-step approach to design new transmembrane pores. We first verified our design in the soluble form because it is much easier to express and characterize proteins when they are not in a lipid bilayer. Then we re-surfaced the pore for membrane association.”

The designed pores are formed from monomeric subunits composed of an inner and an outer helix bridged via short loops. Backbones were generated parametrically by sampling coiled-coil alpha-helical and superhelical Crick parameters. To further refine the structures, hydrogen-bond networks that span the intermolecular interfaces were designed into the pores using Rosetta HBNet. Combinatorial sequence optimization was then used to assign amino acid identities to the remaining residue positions, keeping the polar networks found by HBNet fixed.
Crystal structures of the water-soluble forms of a 12-helical pore and a 16-helical pore closely match the computational design models. Patch-clamp electrophysiology experiments show that, when expressed in insect cells, the transmembrane form of the 12-helix pore enables the passage of ions across the membrane with high selectivity for potassium over sodium; ion passage is blocked by specific chemical modification at the pore entrance. When incorporated into liposomes using in vitro protein synthesis, the transmembrane form of the 16-helix pore—but not the 12-helix pore—enables the passage of biotinylated Alexa Fluor 488. A cryo-electron microscopy structure of the 16-helix transmembrane pore closely matches the design model.
Expression of the largest transmembrane nanopore in insect cells resulted in cell death, probably because of induced cell permeability.
This advance in designing structurally well-defined transmembrane pores, like advances in protein design generally, both inform our understanding of general principles of protein biophysics and open the door for a wide range of applications. “These de novo pores tend to be very stable, so they could be a very good platform for understanding ion selectivity, especially potassium ion selectivity,” said Xu.
This work was supported by the Howard Hughes Medical Institute, National Science Foundation, United States Department of Defense, Air Force Office of Scientific Research, National Science Foundation of China, National Natural Science Foundation of China-Yunnan Joint Fund, Wellcome Trust, Japan Society for the Promotion of Science, and others.