Category: Uncategorized
-
Silk, Wool, and Beyond: AI-Driven Design of Custom Protein Fibers
Today we report in Nature Chemistry a novel approach to designing protein fibers that takes inspiration from silk, wool, and spider webs. This research opens up new avenues in textiles, bioengineering, and beyond. “Silk, wool, and spider webs are all examples of natural protein fibers, and each has a unique blend of flexibility, strength, and…
-
Switch-like proteins inspired by transistors
Today we report in Science the design of custom proteins that can switch between two fully structured conformations. By coupling this molecular motion with a binding event, these proteins serve as biological counterparts to electronic transistors, opening the door to new applications in smart therapeutics, environmental sensing, and more. A team led by postdoctoral scholars…
-

RFdiffusion: A generative model for protein design
A team led by scientists in our lab has created a powerful new way to design proteins by combining structure prediction networks and generative diffusion models. The team demonstrated extremely high computational success and experimentally tested hundreds of A.I.-generated proteins, finding that many may be useful as medications, vaccines, or even new nanomaterials. Originally appearing…
-
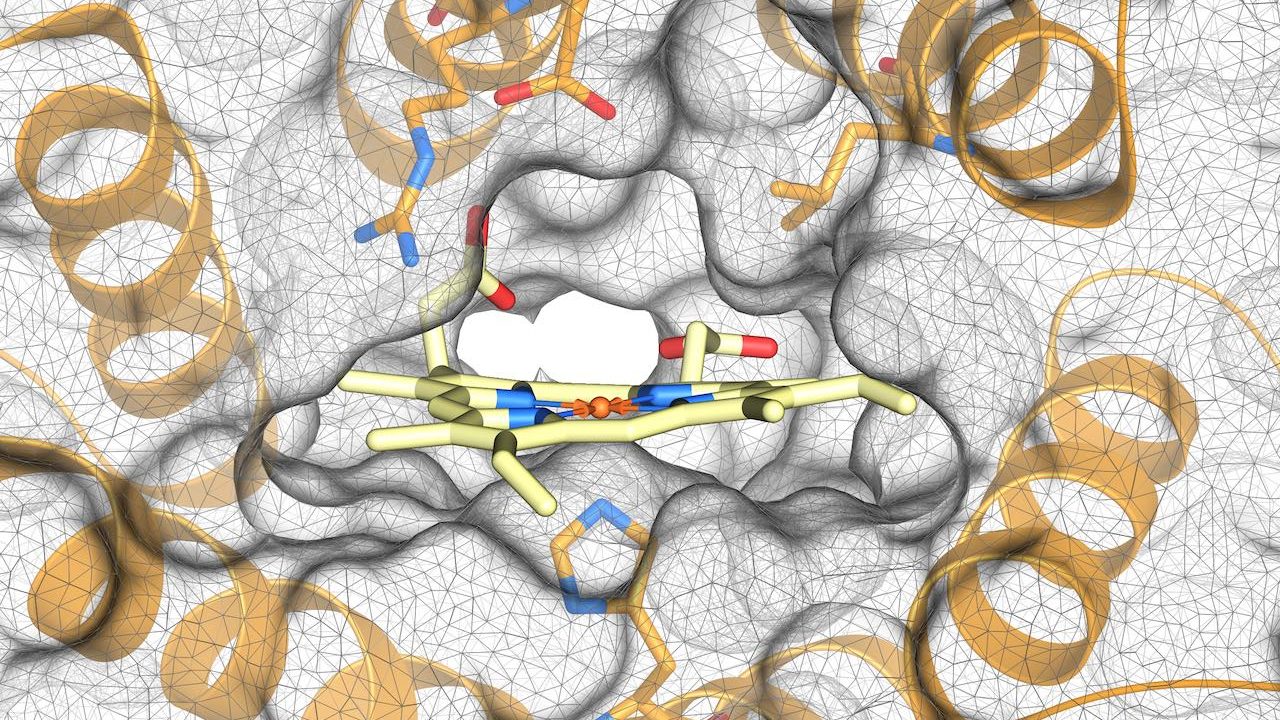
Design of heme-binding enzymes
Today we report in JACS [Open Access] the design of enzymes that contain heme, an important iron-containing molecule involved in many biological processes. The design was inspired by the structure and function of natural heme-binding enzymes, including cytochrome P450, which are involved in a wide range of chemical reactions both inside the body and beyond.…
-

Lab Retreat at Pack Forest
In the beautiful backdrop of the University of Washington’s Pack Forest conference center, our team recently gathered for an immersive lab retreat focused on science and community. As a larger-than-average research group, we believe such gatherings are key to maintaining our culture of collective contribution. They also allow for cross-pollination of ideas among people who…
-
Deep learning improves protein binder design tenfold
In recent years, de novo design of high-affinity protein-binding proteins has become a reality, solely based on target structural information. However, there is still significant room for improvement, as the overall design success rate remains low. In this post, we discuss our lab’s latest open-access research paper published in Nature Communications, which explores the augmentation…
-

Top-down design of protein architectures with reinforcement learning
Today we report in Science [PDF] the successful application of reinforcement learning to a challenge in protein design. This research is a milestone in the use of artificial intelligence for science, and the potential applications are vast, from developing more effective cancer treatments to new biodegradable textiles. A team led by Isaac Lutz, Shunzhi Wang, PhD, and…
-

Design of binders for disordered targets
Today we report in Nature the design of proteins that recognize and bind to the so-called “intrinsically disordered regions” of proteins and peptides. The body produces such disordered molecules naturally, but many have been linked to health disorders, including myeloma and other cancers. “Disordered proteins play important roles in biology. By designing new proteins that…