Today we report the de novo design of protein assemblies that undergo extensive yet precise conformational changes under allosteric control. This early research may lead to advanced tools for precision medicine, vaccine development, and other industrial uses.
This project was led by postdoctoral scholar Arvind Pillai, PhD, and graduate student Abbas Idris, with essential contributions from the Core R&D Labs at the Institute for Protein Design.
What is allostery?
In biology, protein functions are toggled on and off in sophisticated ways. One key mechanism, called allosteric regulation, links functional changes to binding events that occur elsewhere in the protein. This biochemical “action at a distance” is crucial for healthy metabolism and cell signaling, but creating allostery in synthetic protein systems has been a significant challenge — until now.
Designed to move
In a study published today in Nature, we show that RFdiffusion, ProteinMPNN, and other design tools can be used to create an array of dynamic and allosterically switchable protein assemblies. By combining two-state hinges and custom protein-protein interaction modules, we generated assemblies that bear little or no resemblance to any seen before, expanding the possibilities for synthetic biology.
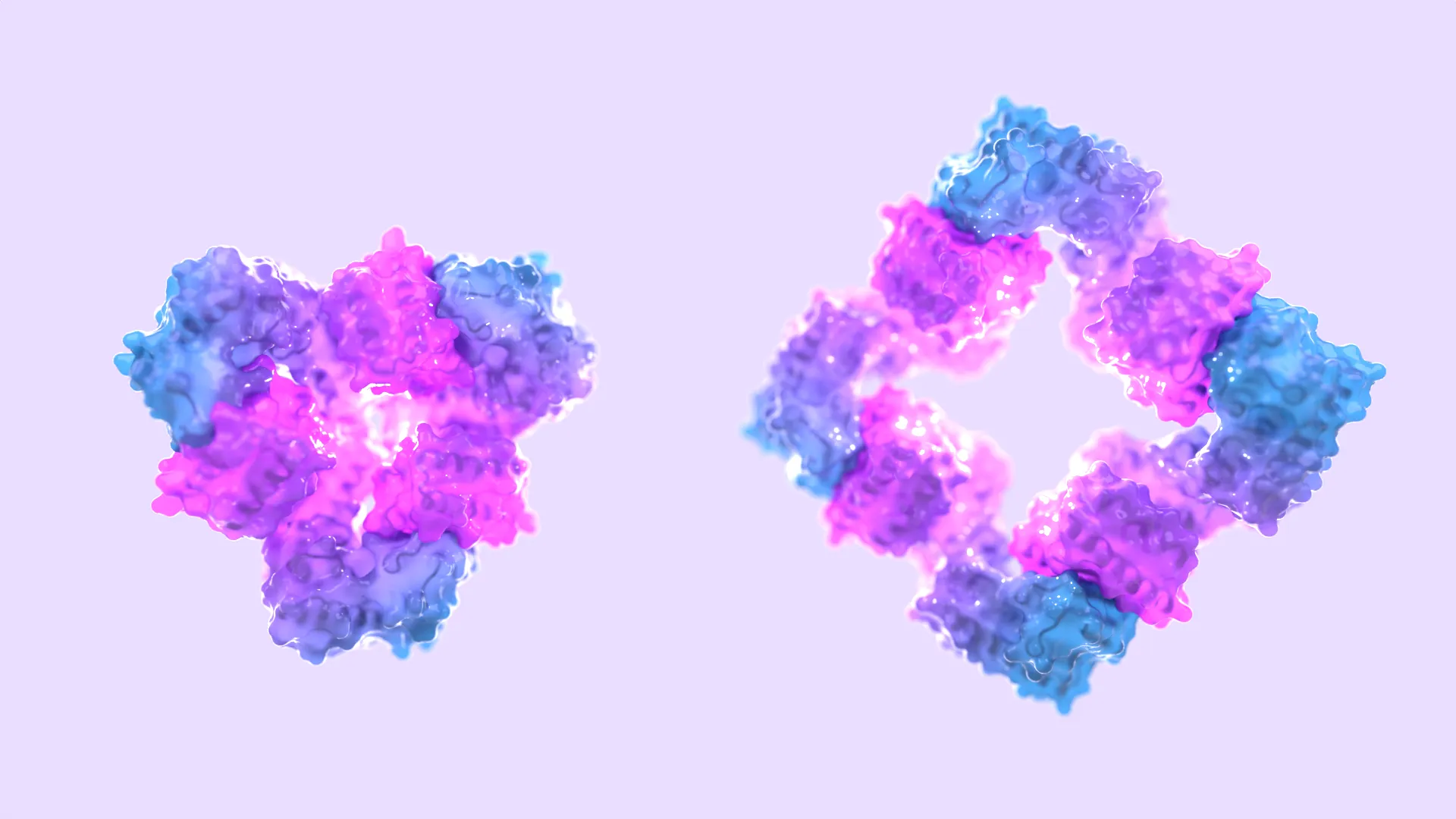
“One of the key innovations in this study is the design of protein assemblies that can switch between different oligomeric states, such as dimers, rings, and cages, in response to effector molecules,” said Arvind. “This ability to control protein structures remotely opens up possibilities for developing adaptive biomaterials and drug delivery systems.”
In addition to structural versatility, the team achieved high-affinity binding between the new proteins and their effectors, ensuring reliable programmed allosteric control. “For this project, we used specific peptides as effectors, but any type of molecule could give rise to protein allostery under the right conditions,” added Abbas.
Remote Control
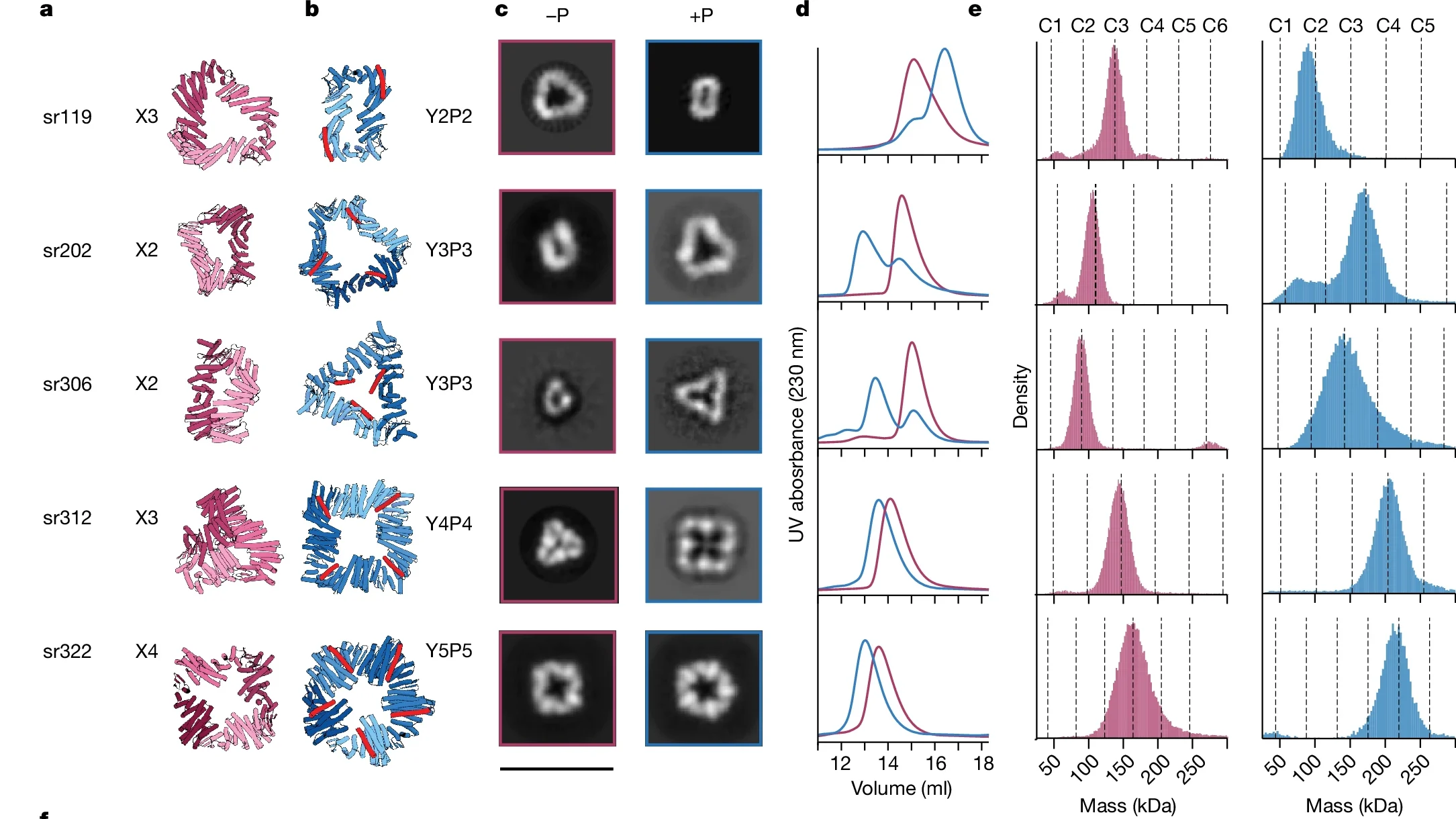
To verify our designs, we characterized more than 20 protein assemblies using a combination of negative stain and cryo-electron microscopy. “This allowed us to confirm which designs formed as intended and to observe how these assemblies altered their structure when effector molecules were introduced,” explained Andrew Borst, PhD, who leads the IPD’s Electron Microscopy Research Core.
We observed allosteric coupling between effector binding sites and assembly interfaces at distances greater than one nanometer—an enormous span in atomic terms. Such extensive coupling is crucial for creating complex protein behaviors that mimic, and potentially surpass, those found in nature.
Potential Applications
The designed assemblies include nano-sized containers that open and close remotely. Such systems may lead to new drug delivery vehicles with advanced control mechanisms, including devices that sequester cell-killing medicines until they encounter a tumor.
This research marks a significant advance in synthetic biology, offering new avenues for creating proteins with precise and controllable functions.
De novo design of allosterically switchable protein assemblies
Published in: Nature [Open Access]
Authors: Arvind Pillai, Abbas Idris, Annika Philomin, Connor Weidle, Rebecca Skotheim, Philip J. Y. Leung, Adam Broerman, Cullen Demakis, Andrew J. Borst, Florian Praetorius, David Baker.


Coverage:
How to design a protein that can be switched on and off — Nature News and Views
AI Designed Proteins Morph on Demand for Steerable Functionality — Genetic Engineering & Biotechnology News
‘Startling Advance’ in Designer Proteins Opens a World of Possibility for Biotech — Singularity Hub