In a new Nature Structure and Molecular Biology study we report the design of custom proteins that can package, protect, and deliver molecular cargo into target cells. This novel drug delivery technology, combined with integrated antibody targeting, opens new applications for precision medicine, potentially allowing medications to be delivered to just the right tissues in the body, thereby reducing side effects and enhancing treatment outcomes for a wide range of diseases.
This project was led by Erin Yang, a recent graduate student in the Baker and King Labs, and included collaborators from the Core R&D Labs at the Institute for Protein Design.

(Re)designing proteins for drug delivery
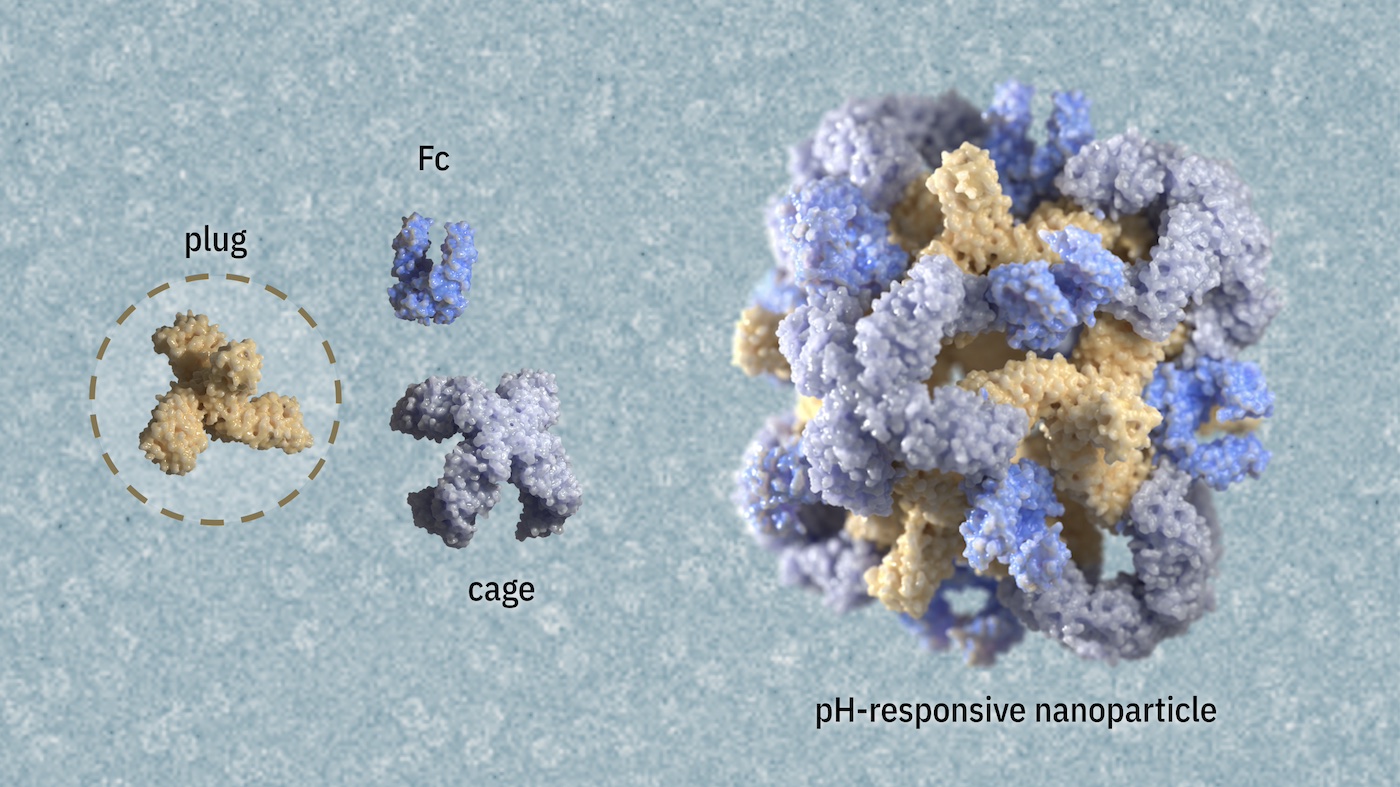
The team began by modifying our previously reported protein nanoparticles that contain integrated antibody domains (Divine et al., 2021). While effective at targeting cells, these nanoparticles had a limitation: their large pores prevent molecular cargo from being stored inside. Our solution? Designing protein “plugs” to transform these nanoparticles into efficient drug delivery vehicles capable of transporting therapeutic proteins, small molecules, or even CRISPR components directly into living cells.
The team used multiple design methods to generate proteins that could plug the pores of existing protein nanoparticles. They confirmed that the desired plugged nanoparticles form in vitro when the three separately purified protein building blocks are combined. Electron microscopists in the IPD Core R&D Labs helped determine the structure of these new nanoparticles.
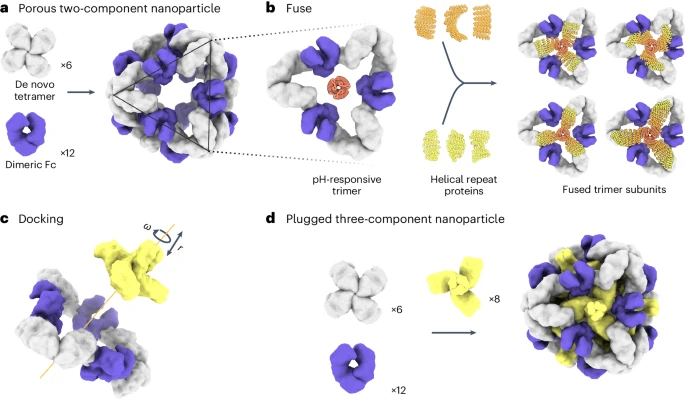
The team also showed that the nanoparticle’s structure remained intact even after one of the protein building blocks — a small antibody-Fc fragment — was swapped out for a larger full-sized antibody. This ability to replace protein parts in a modular fashion should enable a wide range of biosensing and targeting applications.
Responsive Design for Precise Release
Creating proteins that respond to changes in their chemical environment, such as increasing or decreasing acidity, required new design approaches. In the past, most of our designed protein nanoparticles have contained only one or two unique protein building blocks. For this project, however, we saw an opportunity to add a third type, one that could enable pH-responsive disassembly (Boyken et al., 2019).
The team then used Rosetta to tune the amino-acid sequences of the plug proteins to improve pH responsiveness. Particles enter cells through acid-filled compartments called endosomes, which have an internal pH between 4.5 and 6.5. The team designed a suite of nanoparticles that disassembled at pH 5.5 and up to pH 6.8, a range of acidity observed around tumors. In one experiment, the team showed that fluorescent proteins trapped inside the nanoparticles could be released by shifting the chemical environment around the nanoparticles from neutral to acidic.
Laboratory tests on living cells revealed that these nanoparticles show remarkable precision. They can enter human cells displaying target receptors and avoid cells without these receptors, confirming their targeting capabilities.
The development of protein nanoparticles that can encapsulate, deliver, and release different types of molecular cargo inside specific cells is a milestone in the design of new protein functions. The next steps for this project include improving the packaging efficiency of different types of cargo and measuring the efficiency of drug delivery to tumors in animal models.
Funding
Several organizations supported this research. All funders are listed in the manuscript.
Computational design of non-porous pH-responsive antibody nanoparticles
Authors: Erin C. Yang, Robby Divine, Marcos C. Miranda, Andrew J. Borst, Will Sheffler, Jason Z. Zhang, Justin Decarreau, Amijai Saragovi, Mohamad Abedi, Nicolas Goldbach, Maggie Ahlrichs, Craig Dobbins, Alexis Hand, Suna Cheng, Mila Lamb, Paul M. Levine, Sidney Chan, Rebecca Skotheim, Jorge Fallas, George Ueda, Joshua Lubner, Masaharu Somiya, Alena Khmelinskaia, Neil P. King, and David Baker
Published in: Nature Structure and Molecular Biology [PDF]