In a new paper in Nature we introduce a toolkit of standardized protein building blocks that simplify the construction of biomaterials. The approach mirrors the simplicity and programmability seen in conventional construction, where consistent parts allow builders to create structures according to their needs.

This project was led by lab members Timothy Huddy, Yang Hsia, Ryan Kibler, and Jinwei Xu. It also included collaborators from the NYU School of Medicine, M.S. Ramaiah University of Applied Sciences, Lawrence Berkeley National Laboratory, and the Core R&D Labs here at the Institute for Protein Design.
“I took inspiration from my background as a machinist to set a goal of making a style of protein design that is more focused on numbers and dimensions,” said Tim. “We’re already exploring how to construct much larger materials this way, including new protein fibers inspired by spider silk. Other applications we’re exploring include new biomaterials that incorporate minerals like we see with bones and shells, or simplifying the design of vaccines and other molecules for cell biology research.”
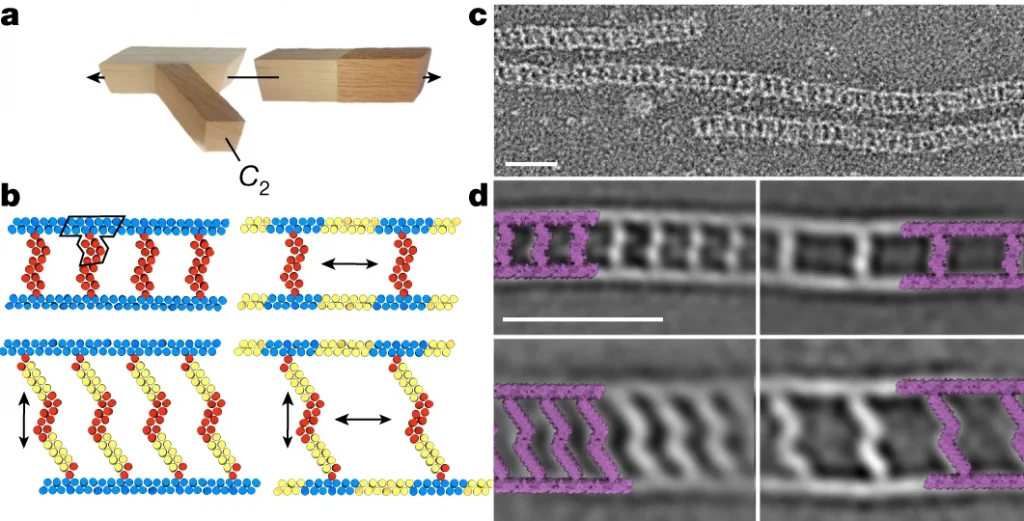
In the study, we describe the development of a new class of proteins that are analogous to perfectly straight, twist-free pieces of lumber. Designed with the aid of AI and then produced in the lab, these parts come in linear, curved, and angled forms. They can be attached together or made longer to create a wide range of intricate shapes.
The team demonstrated this by building several novel biomaterials, each thousands of times thinner than a human hair. This included porous spheres of various sizes and narrow fibers that resemble microscopic railroad tracks.
This work was supported by several funders, including The Audacious Project, National Science Foundation, Open Philanthropy, Howard Hughes Medical Institute, and Institute for Protein Design Breakthrough Fund. A full list of supports is in the paper.
Blueprinting extendable nanomaterials with standardized protein blocks
Authors: Timothy F. Huddy, Yang Hsia, Ryan D. Kibler, Jinwei Xu, Neville Bethel, Deepesh Nagarajan, Rachel Redler, Philip J. Y. Leung, Connor Weidle, Alexis Courbet, Erin C. Yang, Asim K. Bera, Nicolas Coudray, S. John Calise, Fatima A. Davila-Hernandez, Hannah L. Han, Kenneth D. Carr, Zhe Li, Ryan McHugh, Gabriella Reggiano, Alex Kang, Banumathi Sankaran, Miles S. Dickinson, Brian Coventry, T. J. Brunette, Yulai Liu, Justas Dauparas, Andrew J. Borst, Damian Ekiert, Justin M. Kollman, Gira Bhabha, and David Baker.
Published in: Nature [Open Access]
Reviewed in: Nature Reviews Materials [Free Access Link]