Tinberg, C.E., Khare, S.D., et al. Nature. 501(7466), 212-216. (2013)
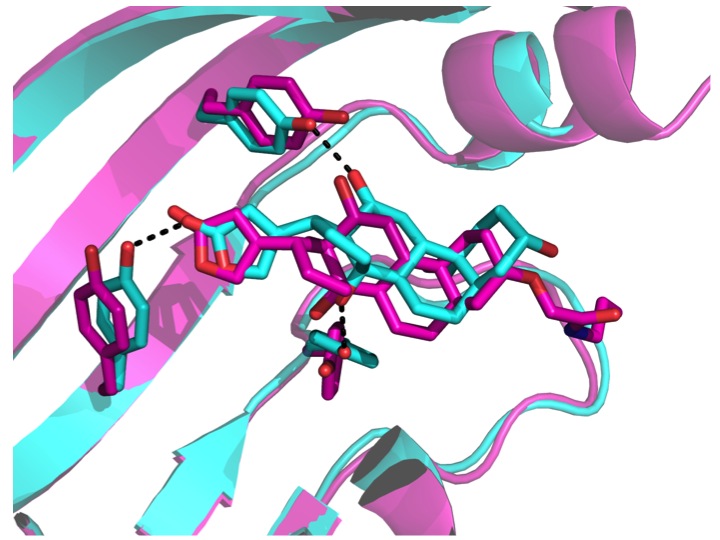
 We describe a computational approach for designing proteins that bind small molecules and use it to create binders for digoxin, a steroid toxin deriving from the foxglove plant. The method relies on the explicit design of highly energetically favorable interactions with the ligand. Directed laboratory evolution guided by deep mutational sequencing can be used to tailor the binding affinity of the designed protein towards the application of interest. The computational method presented here should enable the development of a new generation of biosensors and diagnostics for the detection of small molecule compounds as well as therapeutic proteins for the treatment of small molecule toxicity.
We describe a computational approach for designing proteins that bind small molecules and use it to create binders for digoxin, a steroid toxin deriving from the foxglove plant. The method relies on the explicit design of highly energetically favorable interactions with the ligand. Directed laboratory evolution guided by deep mutational sequencing can be used to tailor the binding affinity of the designed protein towards the application of interest. The computational method presented here should enable the development of a new generation of biosensors and diagnostics for the detection of small molecule compounds as well as therapeutic proteins for the treatment of small molecule toxicity.