Nature (2012)
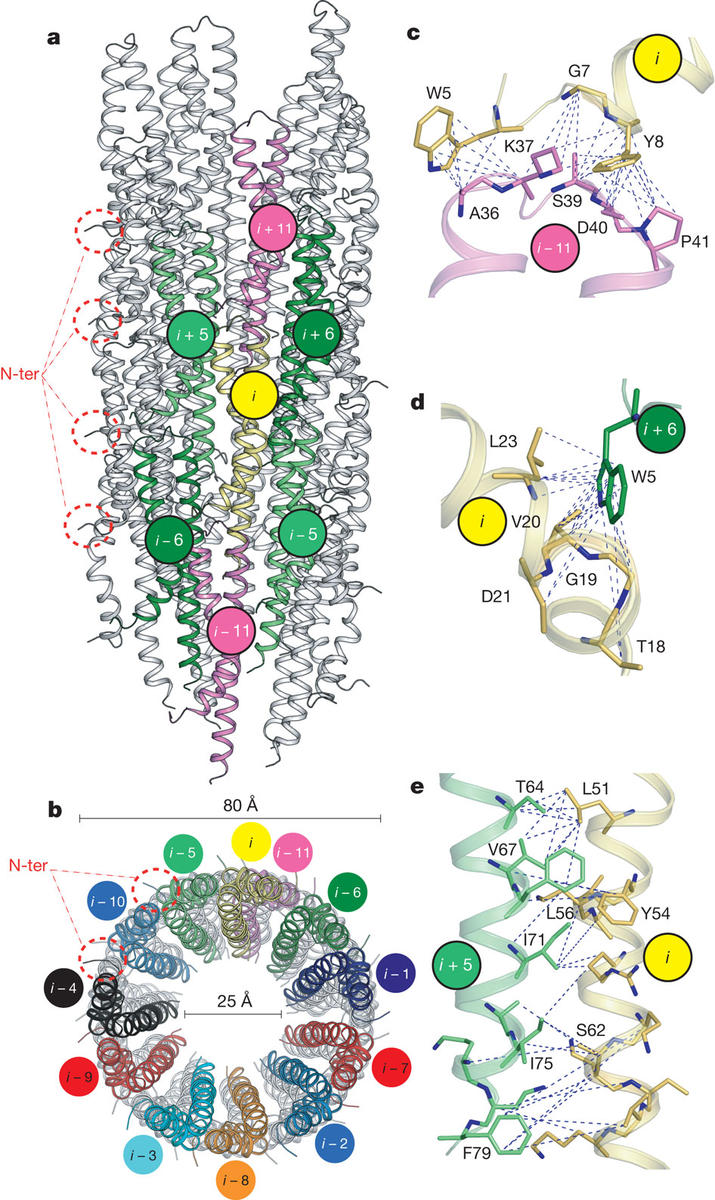
 The ability of Gram-negative bacteria, such as the agents of plague, dysentery and typhoid fever to infect host cells is dependent on a syringe-like molecular machine known as the Type-III secretion system (T3SS). The core of T3SS consists of a hollow filament, the needle; composed of identical, symmetric repeats of an 80-residue protein, the needle forms a conduit for unfolded effector proteins to be delivered to the cytoplasm of the host cell at the early stages of infection. Determination of the three-dimensional structure of the needle by X-ray crystallography or solution NMR has been challenging thus far due to the inherent non-crystallinity and insolubility of the complex. Modeling based on docking of the known monomeric structure into EM reconstructions of isolated needle particles has been limited by the inability of such approaches to capture conformational change as a result of tertiary interactions. We have developed an alternative, hybrid approach through a combination of solid-state NMR data collected in the group of Prof. Adam Lange at the Max Planck Institute, previously published EM data and Rosetta modeling to determine a high-resolution model of in vitro reconstructed needle filaments. We show that the 80-residue subunits form a right-handed helical assembly with roughly 11 subunits per two turns of a 24A-pitch helix. While the more conserved C-terminus is forming key stabilizing towards the inside of the 25A needle pore, the more sequence variant N-terminus is positioned on the surface of the structure. The approach developed here presents a powerful way towards structure determination of large protein assemblies.
The ability of Gram-negative bacteria, such as the agents of plague, dysentery and typhoid fever to infect host cells is dependent on a syringe-like molecular machine known as the Type-III secretion system (T3SS). The core of T3SS consists of a hollow filament, the needle; composed of identical, symmetric repeats of an 80-residue protein, the needle forms a conduit for unfolded effector proteins to be delivered to the cytoplasm of the host cell at the early stages of infection. Determination of the three-dimensional structure of the needle by X-ray crystallography or solution NMR has been challenging thus far due to the inherent non-crystallinity and insolubility of the complex. Modeling based on docking of the known monomeric structure into EM reconstructions of isolated needle particles has been limited by the inability of such approaches to capture conformational change as a result of tertiary interactions. We have developed an alternative, hybrid approach through a combination of solid-state NMR data collected in the group of Prof. Adam Lange at the Max Planck Institute, previously published EM data and Rosetta modeling to determine a high-resolution model of in vitro reconstructed needle filaments. We show that the 80-residue subunits form a right-handed helical assembly with roughly 11 subunits per two turns of a 24A-pitch helix. While the more conserved C-terminus is forming key stabilizing towards the inside of the 25A needle pore, the more sequence variant N-terminus is positioned on the surface of the structure. The approach developed here presents a powerful way towards structure determination of large protein assemblies.