Tag: hydrogel
-
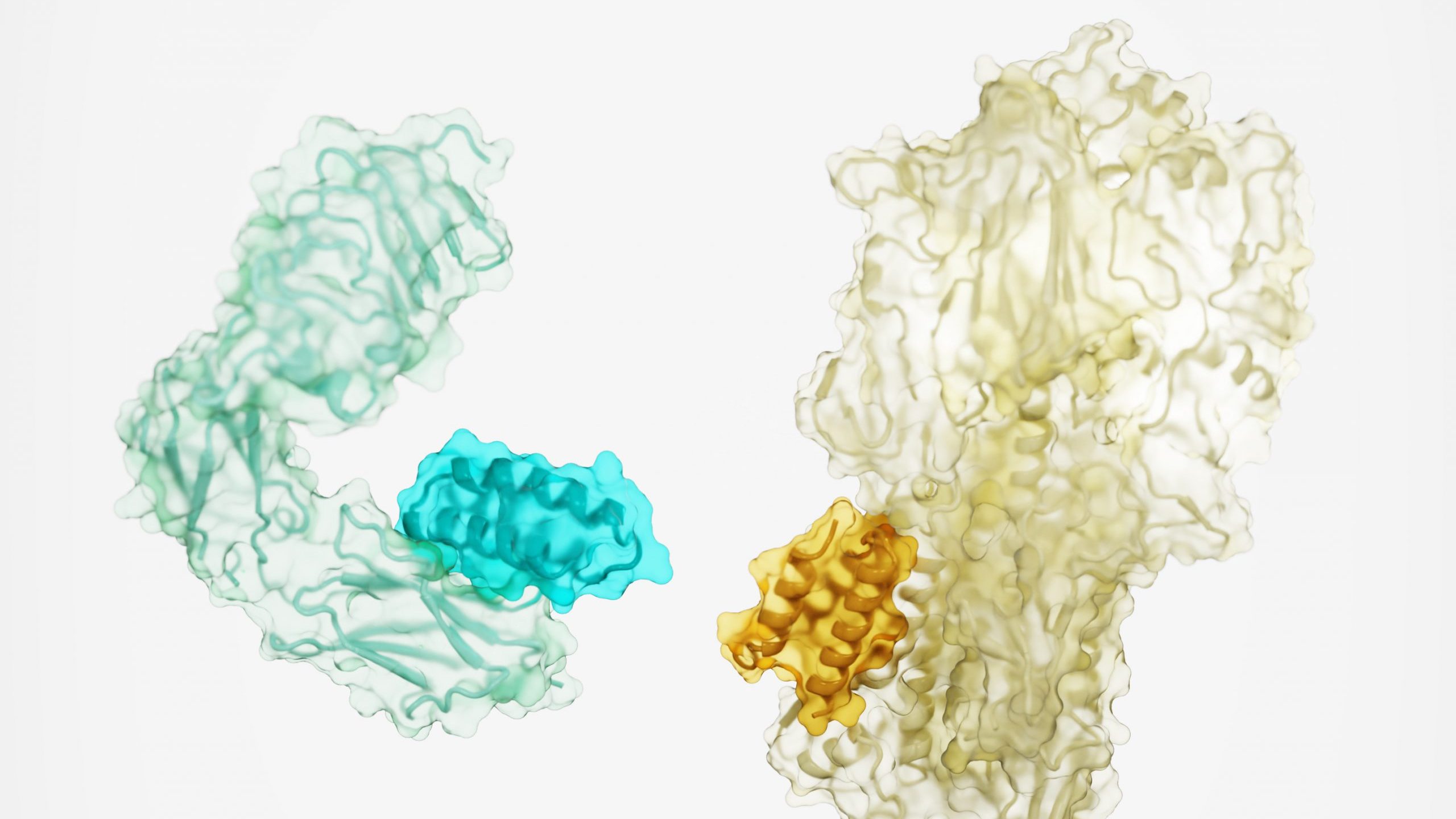
Design of proteins binders from target structure alone
Today we report in Nature a new method for generating protein drugs. Using Rosetta-based design, an international team designed molecules that can target important proteins in the body, such as the insulin receptor, as well as proteins on the surface of viruses. This solves a long-standing challenge in drug development and may lead to new…
-

Exploring new landscapes
-

Watch David Baker speak at the PDB 50 Year Anniversary
The Protein Data Bank (PDB), an archive of free macromolecular structural data, recently marked its 50th birthday. To celebrate, Biophysical Society hosted a virtual symposium, highlighting some of the high-impact applications of protein structural data, with a particular focus on the areas of structure prediction and membrane protein biophysics. At the event, David Baker shared…
-

Diverse protein assemblies by (negative) design
A new approach for creating custom protein complexes yields asymmetric assemblies with interchangeable parts. Today we report in Science the design of new protein assemblies made from modular parts. These complexes — which adopt linear, branching, or closed-loop architectures — contain up to six unique proteins, each of which remains folded and soluble in the absence…
-

Deep learning dreams up new protein structures
Just as convincing images of cats can be created using artificial intelligence, new proteins can now be made using similar tools. In a new report in Nature, we describe the development of a neural network that “hallucinates” proteins with new, stable structures. “For this project, we made up completely random protein sequences and introduced mutations into…
-
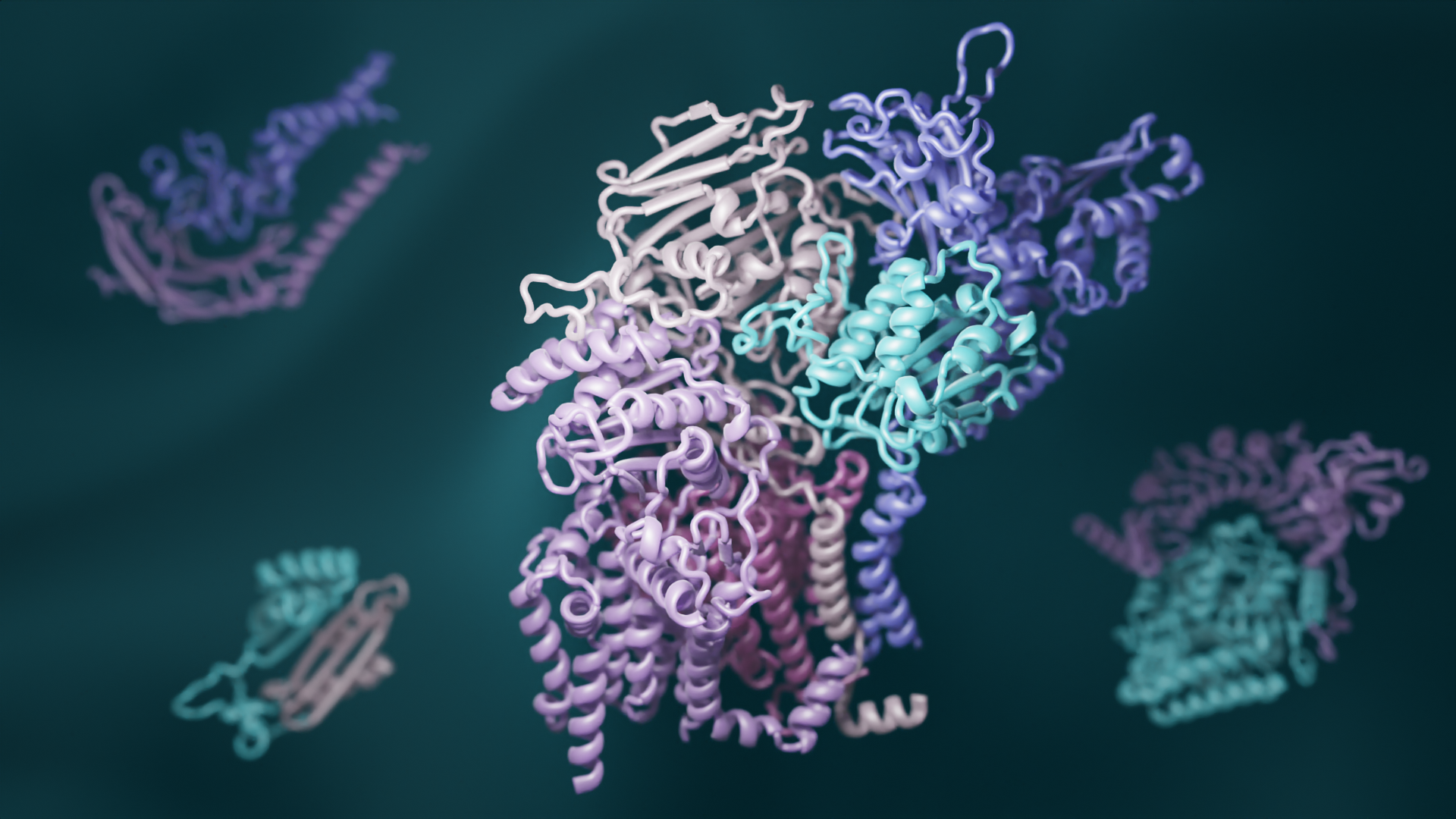
Deep learning reveals how proteins interact
A team led by scientsts in the Baker lab has combined recent advances in evolutionary analysis and deep learning to build three-dimensional models of how most proteins in eukaryotes interact. This breakthrough has significant implications for understanding the biochemical processes common to all animals, plants, and fungi. This open-access work appears in Science. “To really…
-

Lab hike to Lake Ingalls
Several postdocs and grad students from the lab trekked out to the Alpine Lakes Wilderness. Luckily, a few hikers remembered to pack their cameras! [envira-gallery id=”3577″]
-

Accurate protein structure prediction accessible to all
Today we report the development and initial applications of RoseTTAFold, a software tool that uses deep learning to quickly and accurately predict protein structures based on limited information. Without the aid of such software, it can take years of laboratory work to determine the structure of just one protein. With RoseTTAFold, a protein structure can be…