Publications
Design of high- specificity binders for peptide–MHC- I complexes
Published in: Science
Authors: Bingxu Liu, Nathan F. Greenwood, Julia E. Bonzanini, Amir Motmaen, Jeremy Meyerberg, Tao Dao, Xinyu Xiang, Russell Ault, Jazmin Sharp, Chunyu Wang, Gian Marco Visani, Dionne K. Vafeados, Nicole Roullier, Armita Nourmohammad, David A. Scheinberg, K. Christopher Garcia, David Baker
Lead Authors


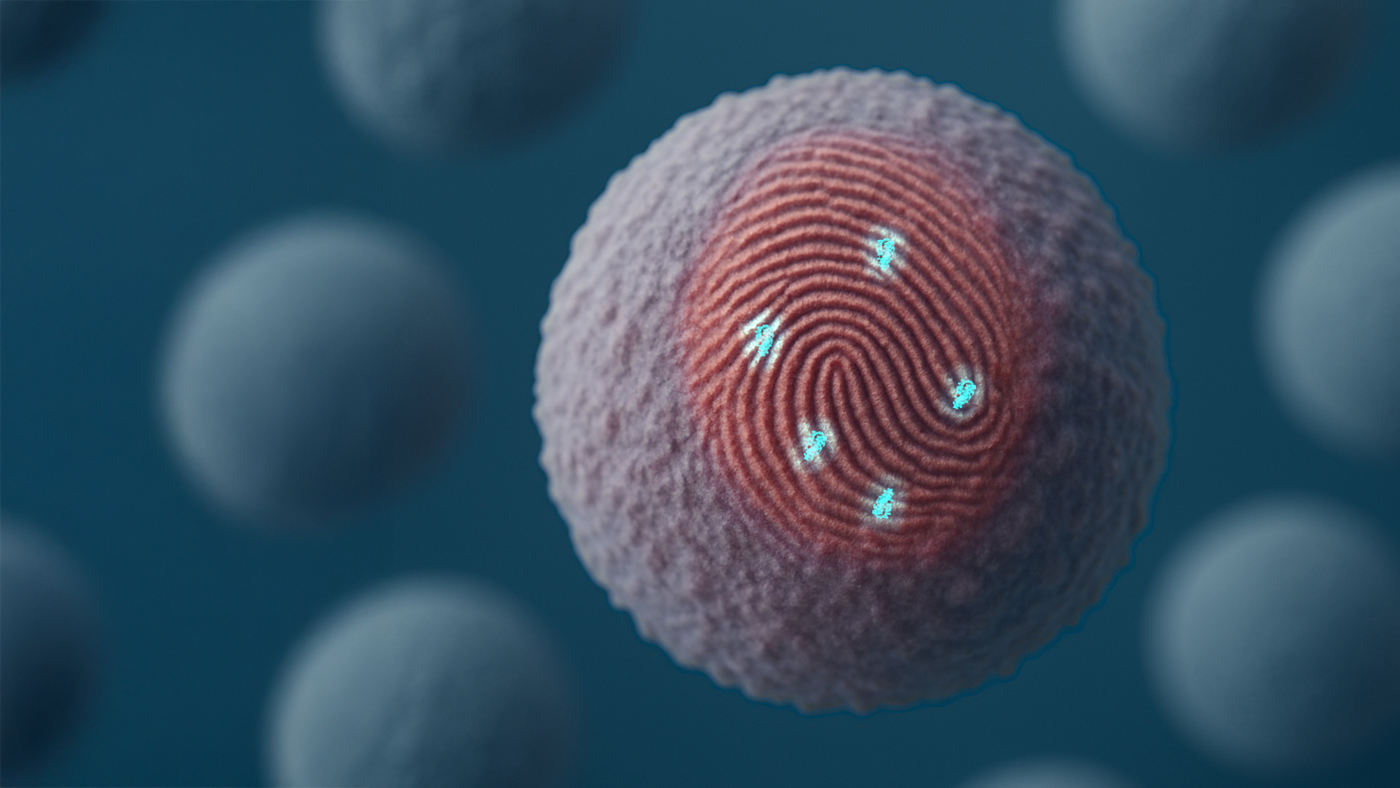
The immune system constantly scans the body for subtle signs of disease. Natural protein systems on the surface of most cells called major histocompatibility complexes (MHCs) support this process by displaying cellular content for immune surveillance. But some disease markers fail to trigger a strong immune response, allowing certain illnesses to persist in the body.
To help overcome this, we have developed the first fully computational approach for enhancing the immune system’s ability to precisely detect and destroy cells bearing more elusive disease markers.
Published today in the journal Science, this research was led by Bingxu Liu, Nathan Forest Greenwood, and Julia Bonzanini with collaborators from the Nourmohammad Lab at the University of Washington, Scheinberg Lab at Memorial Sloan Kettering Cancer Center, Garcia Lab at Stanford University, and Core R&D Labs at the UW Institute for Protein Design.
Building pMHC Binders
A key challenge in targeting peptide-MHCs is recognizing the specific peptides on display while avoiding unintended interactions with MHC proteins themselves. Disease markers may differ from healthy ones by as little as one amino acid substitution, and medicines that mistakenly lock onto MHC proteins directly risk directing immune cells to attack healthy tissues everywhere the body.
Using RFdiffusion and ProteinMPNN, the team designed small proteins that arch over MHC molecules and make highly specific contact with disease-associated peptides. They tested this new binder design approach on 11 different peptide-MHC targets, including fragments from HIV and mutated peptides linked to cancer.
When the designed proteins were incorporated into chimeric antigen receptors (CARs) — components of some cell therapies — immune cell activation was achieved for eight of the targets. Two designed proteins were also shown to activate the targeted killing of human cells displaying disease markers in lab tests.
“Since these proteins are cheap to develop, I’m hopeful this will lead to new therapies that are more accessible to patients worldwide.”
Julia Bonzanini
Binding experiments further revealed that each functional binder only stuck to the target it was built for, and x-ray crystallography of one bound complex confirmed the new binder was built with near-atomic accuracy, validating the design process.
One standout target was the tumor antigen PRAME, which is abundant in many cancers. Because no high-resolution structure existed for PRAME-MHC, the team used AlphaFold3 to predict it, then created functional binders using the simulated target structure. CAR-T cells equipped with these novel PRAME-targeting proteins selectively destroyed cells bearing the PRAME marker without harming similar healthy cells.
Speed and Scalability
The design process also proved highly adaptable. Starting from one successful AI-generated binder, it took the team less than one week to create new versions capable of binding to different tumor and viral peptide targets.
“Because our approach is fully computational, it can be applied to a wide range of disease markers, including those linked to currently untreatable conditions.”
Nathan Greenwood
Unlike traditional drug discovery methods that often rely on screening vast experimental libraries, this approach is entirely digital. Once a target is defined, thousands of potential binders can be generated and evaluated digitally — all before entering the lab. This shrinks development timelines and reduces complexity while opening the door to more personalized medicines.
Next Steps
“We’ve shown how advances in protein design could make personalized cancer therapy possible, and we intend to start a company to turn these results into real therapies that benefit patients.”
Bingxu Liu, PhD
The University of Washington has filed a provisional patent on the proteins developed in this study which have not yet been tested in animals or humans. The protein design software that enabled this project is available online and free for all researchers to use.
Funding
Funding for this study was provided by The Audacious Project, Howard Hughes Medical Institute, Parker Institute for Cancer Immunotherapy, Washington Research Foundation, Children’s Hospital of Philadelphia, Microsoft, Cancer Grand Challenges team MATCHMAKERS (funded by Cancer Research UK, the US National Cancer Institute and The Mark Foundation for Cancer Research), and the US Department of Defense (HDTRA), National Science Foundation (2045054), and National Institutes of Health (2R01AI103867-11, P30 CA008748, R35 GM142795, R35 CA241894, R50 CA265328).