Today we report in Cell on the de novo design of proteins that direct human stem cells to form new blood vessels in the lab. This newfound control over stem cell development is a step toward more effective regenerative medicines.
“Whether through heart attack, diabetes, and the natural process of aging, we all accumulate damage in our body’s tissues. One way to repair some of this damage may be to drive the formation of new blood vessels in areas that need healthy blood supply restored,” said Hannele Ruohola-Baker, a senior author of the study. She is a professor of biochemistry and associate director of the Institute for Stem Cell and Regenerative Medicine at UW Medicine.
Rearranging receptors
Growth factors play key roles in tissue development, wound healing, and cancer. By binding to growth factor receptors on the outside of cells, these molecules drive changes inside. Researchers have for decades attempted to repurpose natural growth factors as regenerative medicines with some limited success, but many of these experimental treatments have failed due to imprecision.
“We set out to create custom proteins that would engage with cellular growth factor receptors in extremely precise ways. When we made these molecules in the lab and treated human stem cells with them, we saw different kinds of vasculature develop depending on which proteins we used. This is a whole new level of control,” explained Natasha Edman, a lead author of the study and recent member of the lab.
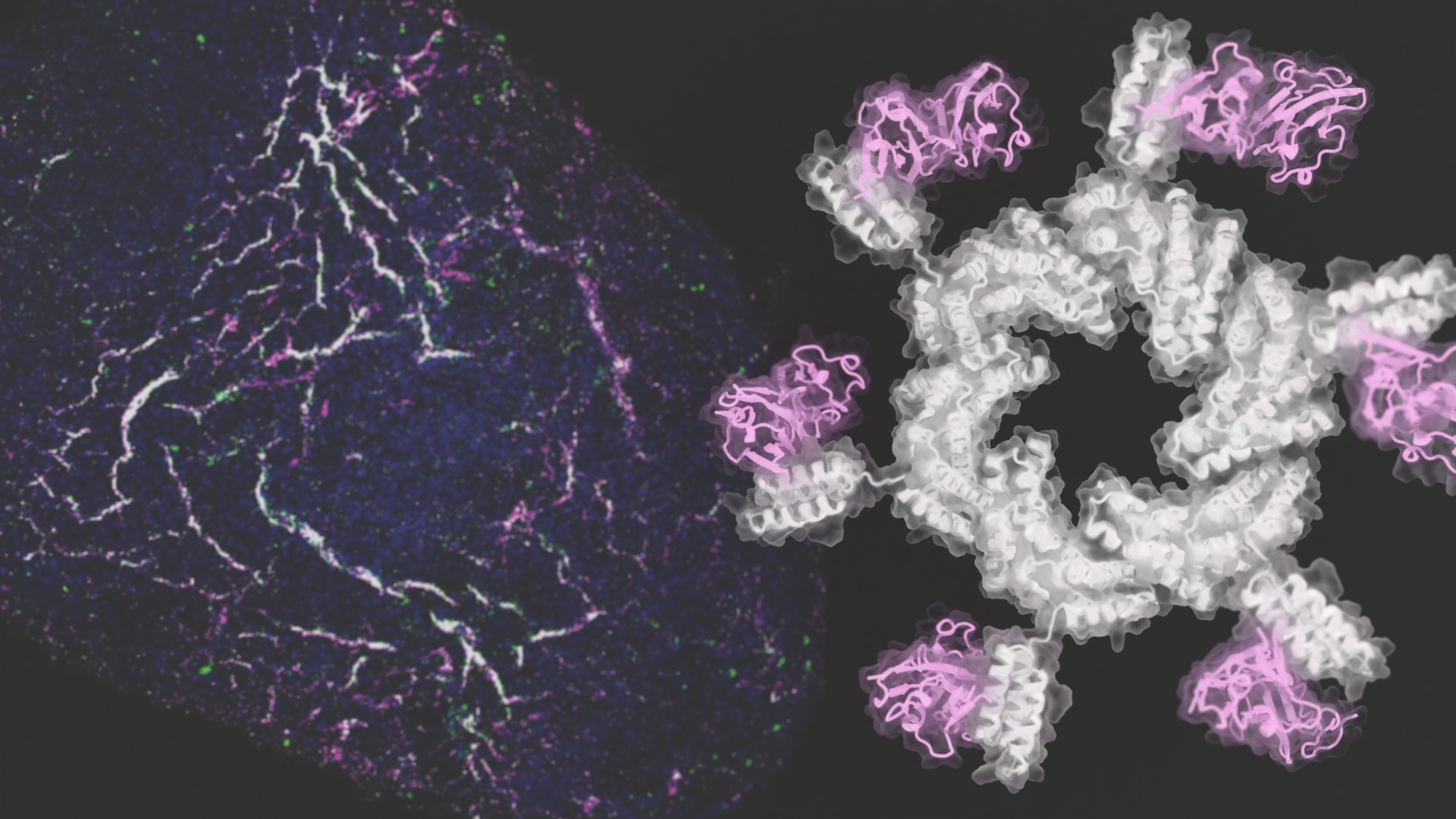
Development by design
The team designed ring-shaped proteins, each targeting up to eight fibroblast growth factor receptors. By varying the size of the rings and other protein properties, they could control how stem cells matured under laboratory conditions.
The resulting vascular networks were functional and mature. They formed tubes, healed when scratched, and absorbed nutrients from their surroundings as expected. When transplanted into mice, these tiny webs of human blood vessels grew connections to the animal’s circulatory system within three weeks.
“We decided to focus on building blood vessels first, but this same technology should work for many other types of tissues. This opens up a new way of studying tissue development and could lead to a new class of medicines for spinal cord injury and other conditions that have no good treatment options today,” said Ashish Phal, a lead study author and bioengineering Ph.D. candidate at UW.
Dr. Ruohola-Baker adds that with this investigation, a code has been cracked. For the first time, designed proteins have been used to direct stem cells to become the endothelial cells that form the walls of arteries, a breakthrough that will help scientists model diseases and regenerate this type of blood vessel.
This research was performed at the Institute for Protein Design and Institute for Stem Cell and Regenerative Medicine at UW Medicine and included collaborators from New York University School of Medicine, Tehran University of Medical Sciences, Yale University School of Medicine, Brotman Baty Institute for Precision Medicine, and Allen Discovery Center for Cell Lineage Tracing.
Funding
This work was supported by The Audacious Project, Open Philanthropy, Nordstrom-Barrier Directors Fund, Institute for Protein Design Breakthrough Fund, Brotman Baty Institute, Institute for Stem Cell and Regenerative Medicine Fellows Program, Howard Hughes Medical Institution (GT11817), American Heart Association (19IPLOI34760143), Human Frontier Science Program (LT000880/2019-L), Simons Foundation (SF349247), New York State Assembly, National Institutes of Health (T90DE021984, R01GM097372, R01GM083867, 1P01GM081619, COF220919, U24 GM129539, P30 GM124169, S10OD018483), National Institute of General Medical Sciences (R35GM128777, R35GM150919), National Institute on Aging (R01AG063845, U19AG065156), National Heart, Lung, and Blood Institute (U01HL099997, UO1HL099993), Department of Defense (PR203328 W81XWH-21-1-0006), Department of Energy, and European Molecular Biology Organization (ALTF191-2021).
Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies
Authors: Natasha I. Edman, Ashish Phal, Rachel L. Redler, Thomas Schlichthaerle, Sanjay R. Srivatsan, Devon Duron Ehnes, Ali Etemadi, Seong J. An, Andrew Favor, Zhe Li, Florian Praetorius, Max Gordon, Thomas Vincent, Silvia Marchiano, Leslie Blakely, Chuwei Lin, Wei Yang, Brian Coventry, Derrick R. Hicks, Longxing Cao, Neville Bethel, Piper Heine, Analisa Murray, Stacey Gerben, Lauren Carter, Marcos Miranda, Babak Negahdari, Sangwon Lee, Cole Trapnell, Ying Zheng, Charles E. Murry, Devin K. Schweppe, Benjamin S. Freedman, Lance Stewart, Damian C. Ekiert, Joseph Schlessinger, Jay Shendure, Gira Bhabha, Hannele Ruohola-Baker, David Baker.
Published in: Cell [Open Access]