Rothlisberger D., Khersonsky O. et al, Nature, 453, 164-6. (2008)
Khersonsky O., Rothlisberger D. et al, J Mol Biol, 396(4), 1025-42. (2010)
 We designed several enzymes that catalyze Kemp elimination, a model reaction for proton transfer from carbon. These enzymes utilize two catalytic motifs and enhance the reaction rate up to 105-fold with multiple turnovers. Application of in vitro evolution to enhance the computational designs KE07, KE70, and KE59 produced up to 2000-fold increases in catalytic activity, with kcat/Km values reaching 105-106 M-1s-1.
We designed several enzymes that catalyze Kemp elimination, a model reaction for proton transfer from carbon. These enzymes utilize two catalytic motifs and enhance the reaction rate up to 105-fold with multiple turnovers. Application of in vitro evolution to enhance the computational designs KE07, KE70, and KE59 produced up to 2000-fold increases in catalytic activity, with kcat/Km values reaching 105-106 M-1s-1.
High-resolution crystal structures of initial designs and their evolved variants demonstrated that the designs have close to atomic accuracy, and provided insights into the origins of the improvements in catalysis.
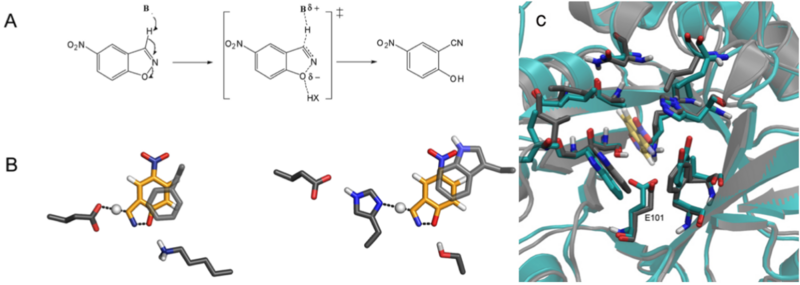
A. The Kemp elimination proceeds by means of a single transition state, which can be stabilized by a base deprotonating the carbon and the dispersion of the resulting negative charge; a hydrogen bond donor can also be used to stabilize the partial negative charge on the phenolic oxygen. B. Examples of active site motifs highlighting the two choices for the catalytic base (a carboxylate (left) or a His–Asp dyad (right)) used for deprotonation, and a p-stacking aromatic residue for transition state stabilization. C. Comparison of the designed model of KE07 and the crystal The crystal structure (cyan) was solved in the unbound state and shows only modest rearrangement of active site side chains compared to the designed structure (grey) modelled in the presence of the transition state (yellow, transparent).